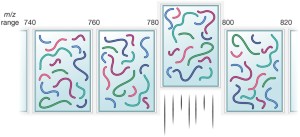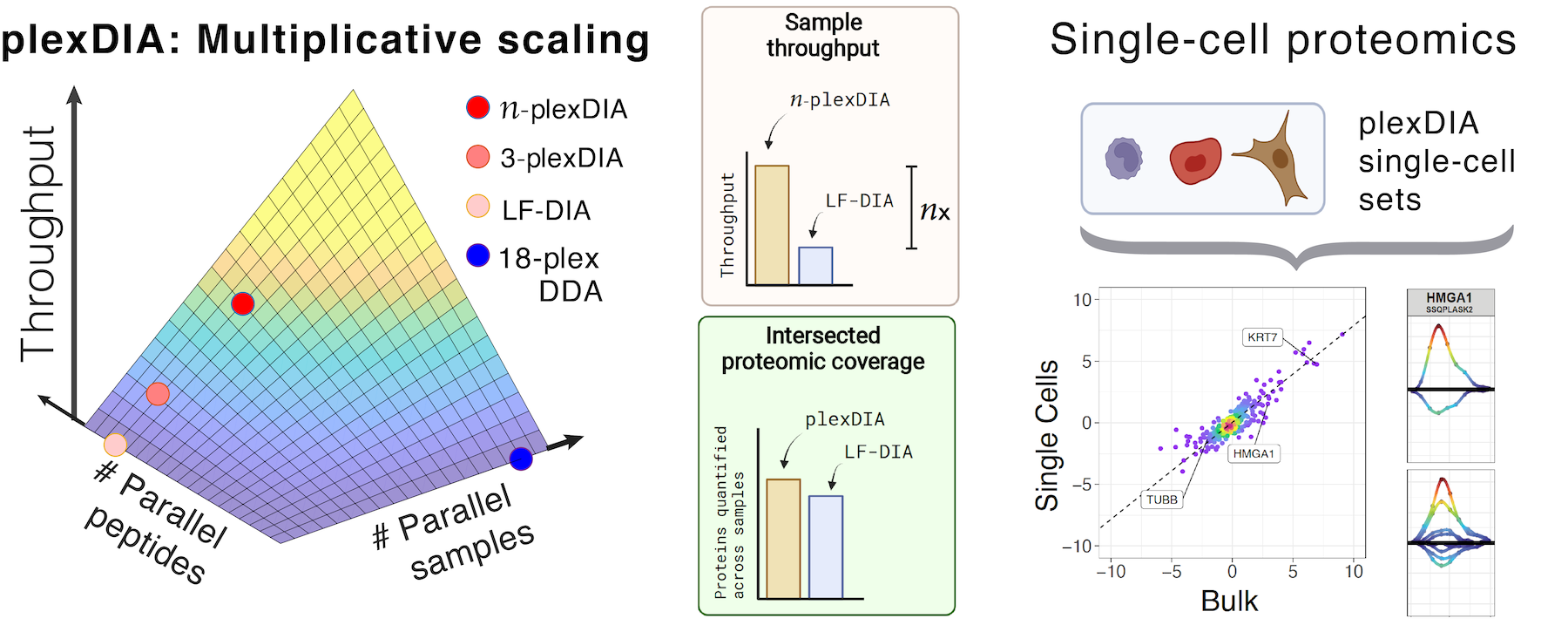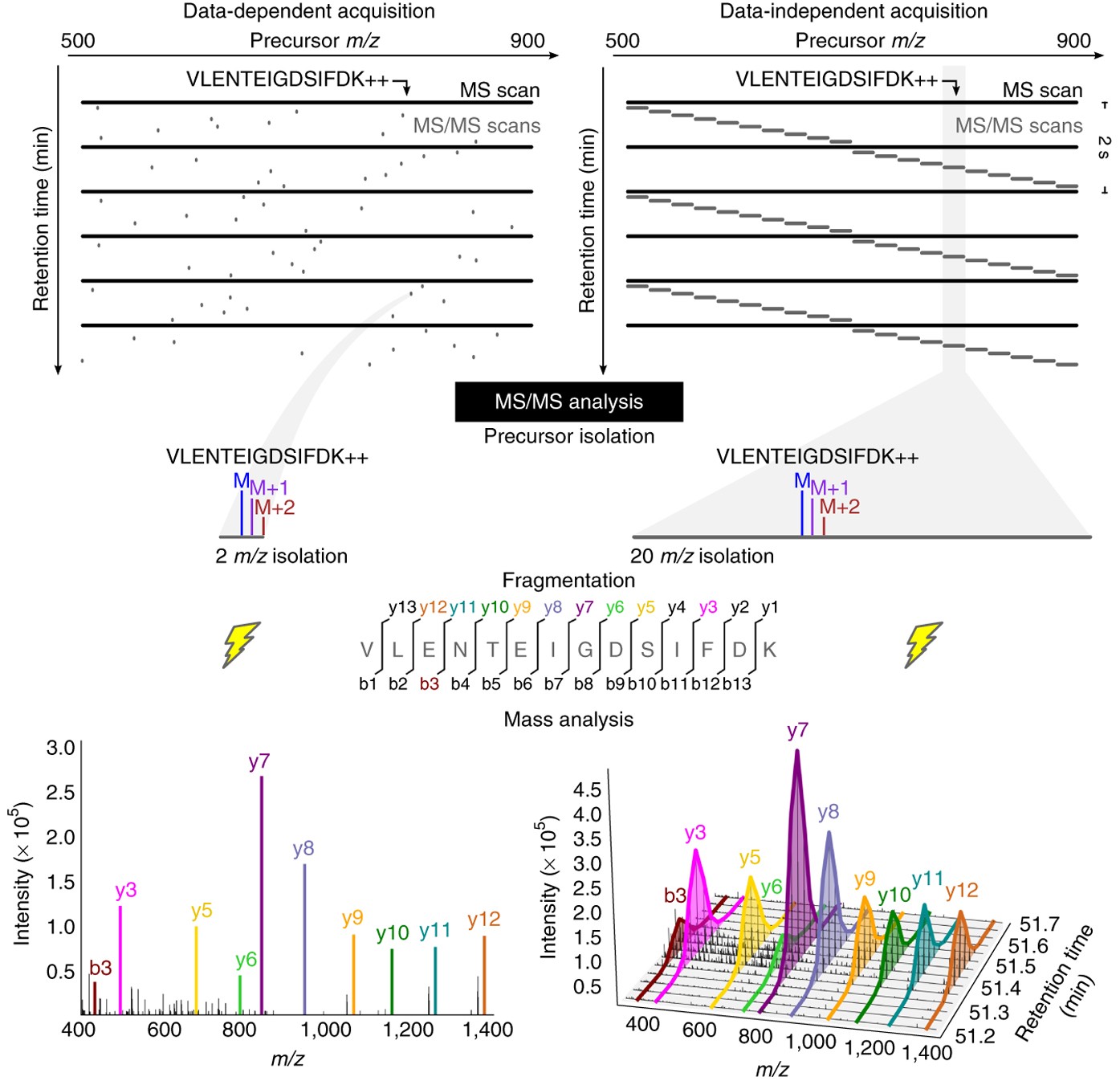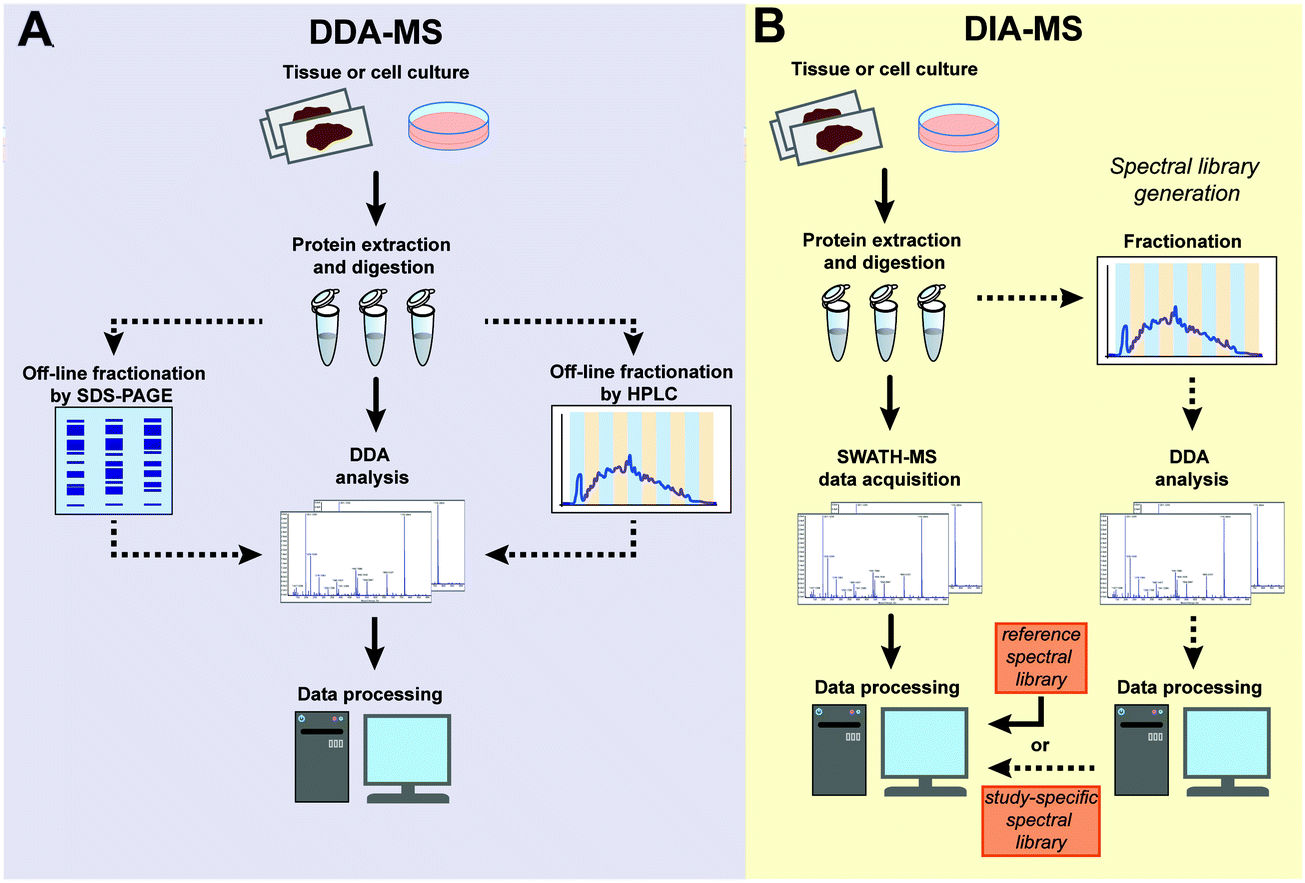
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H
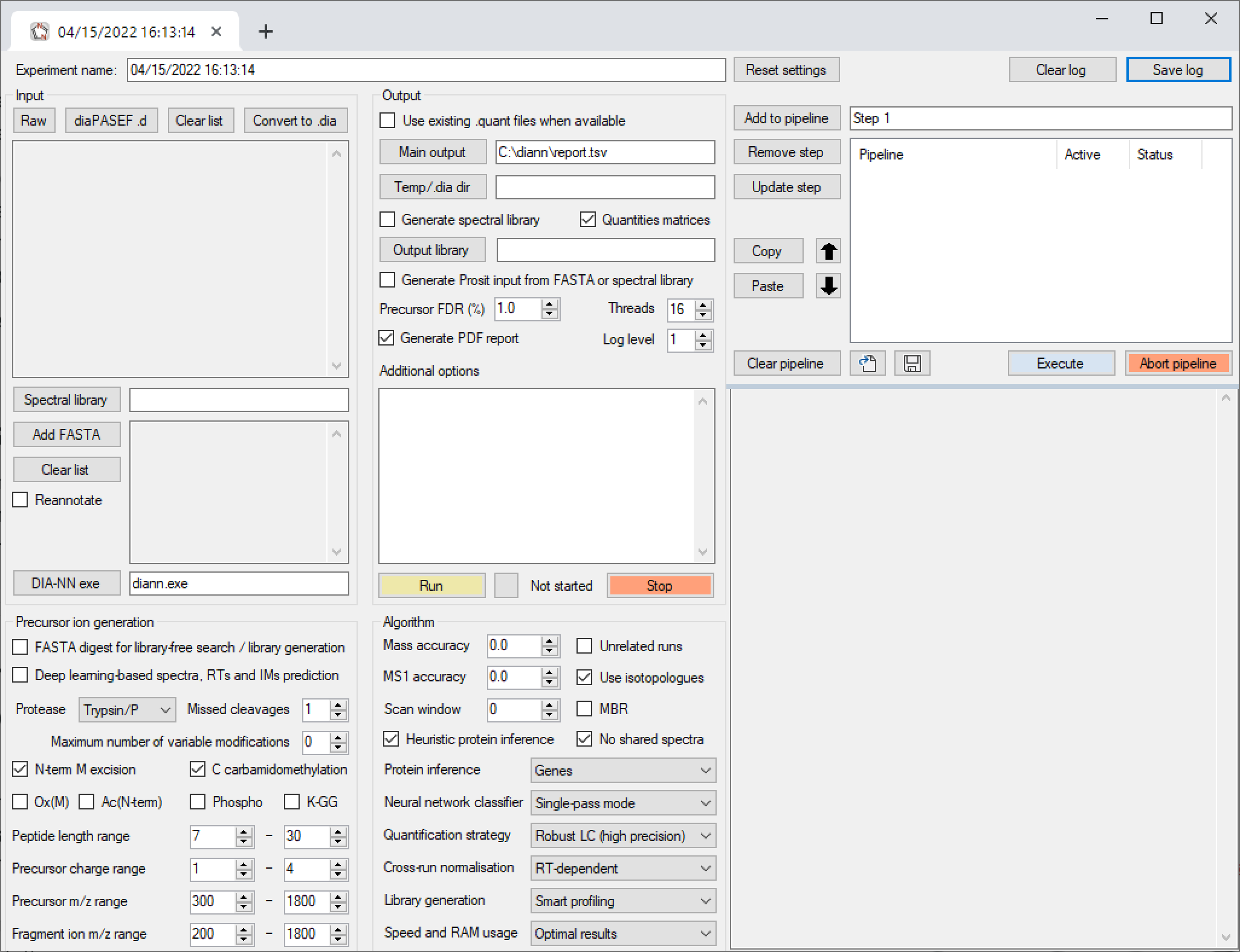
GitHub - vdemichev/DiaNN: DIA-NN - a universal automated software suite for DIA proteomics data analysis.

Variability in Mass Spectrometry-based Quantification of Clinically Relevant Drug Transporters and Drug Metabolizing Enzymes | Molecular Pharmaceutics

DIAproteomics: A Multifunctional Data Analysis Pipeline for Data-Independent Acquisition Proteomics and Peptidomics | Journal of Proteome Research
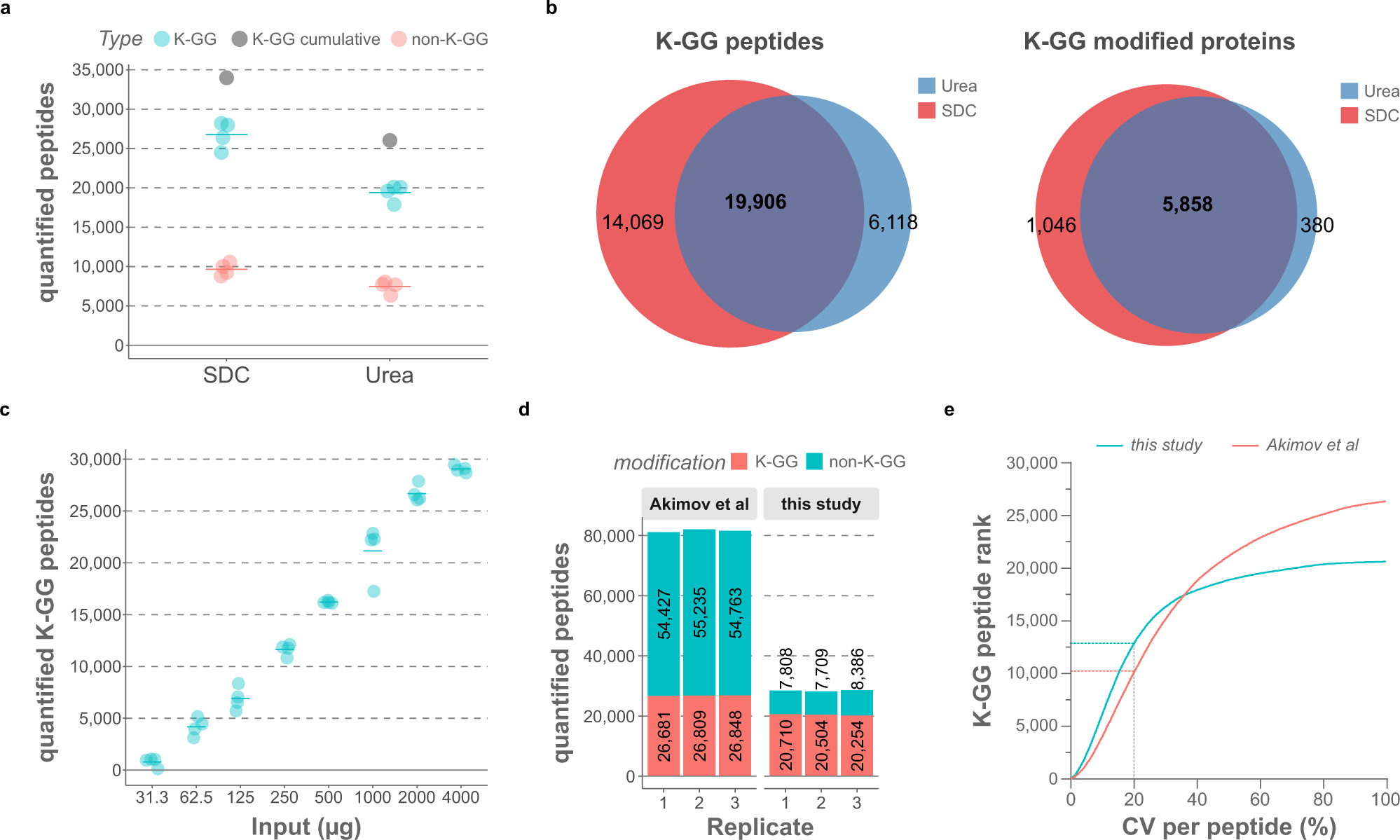
Time-resolved in vivo ubiquitinome profiling by DIA-MS reveals USP7 targets on a proteome-wide scale | Nature Communications

Implementing the reuse of public DIA proteomics datasets: from the PRIDE database to Expression Atlas | Scientific Data
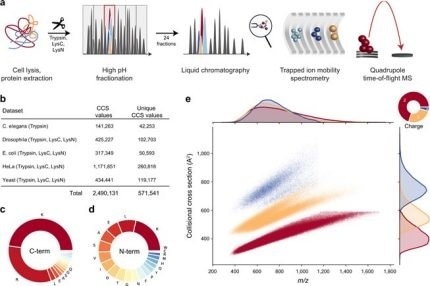
New Nature Communications publication by Mann & Theis Groups harnesses the benefits of large-scale peptide collisional cross section (CCS) measurements and deep learning for 4D-proteomics
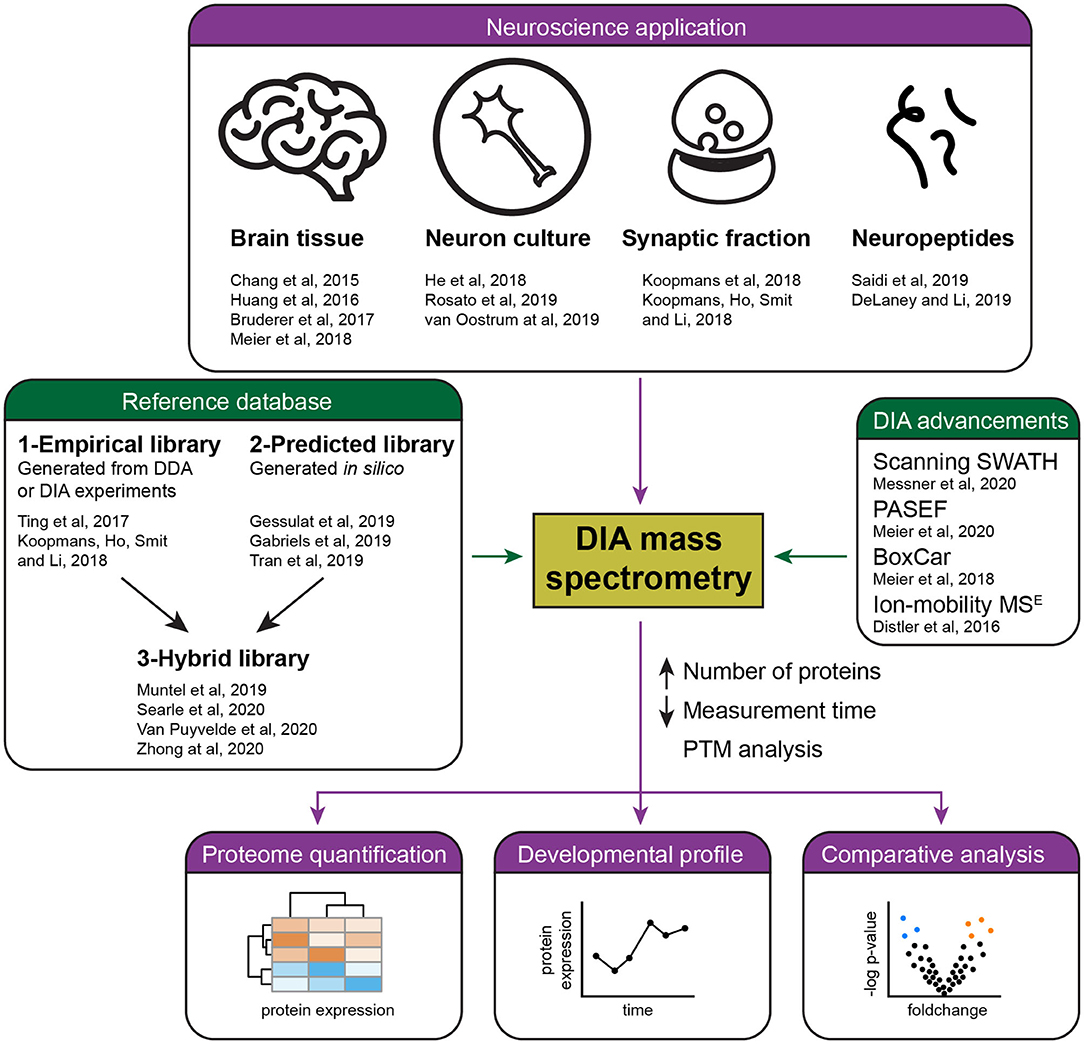
Frontiers | Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome
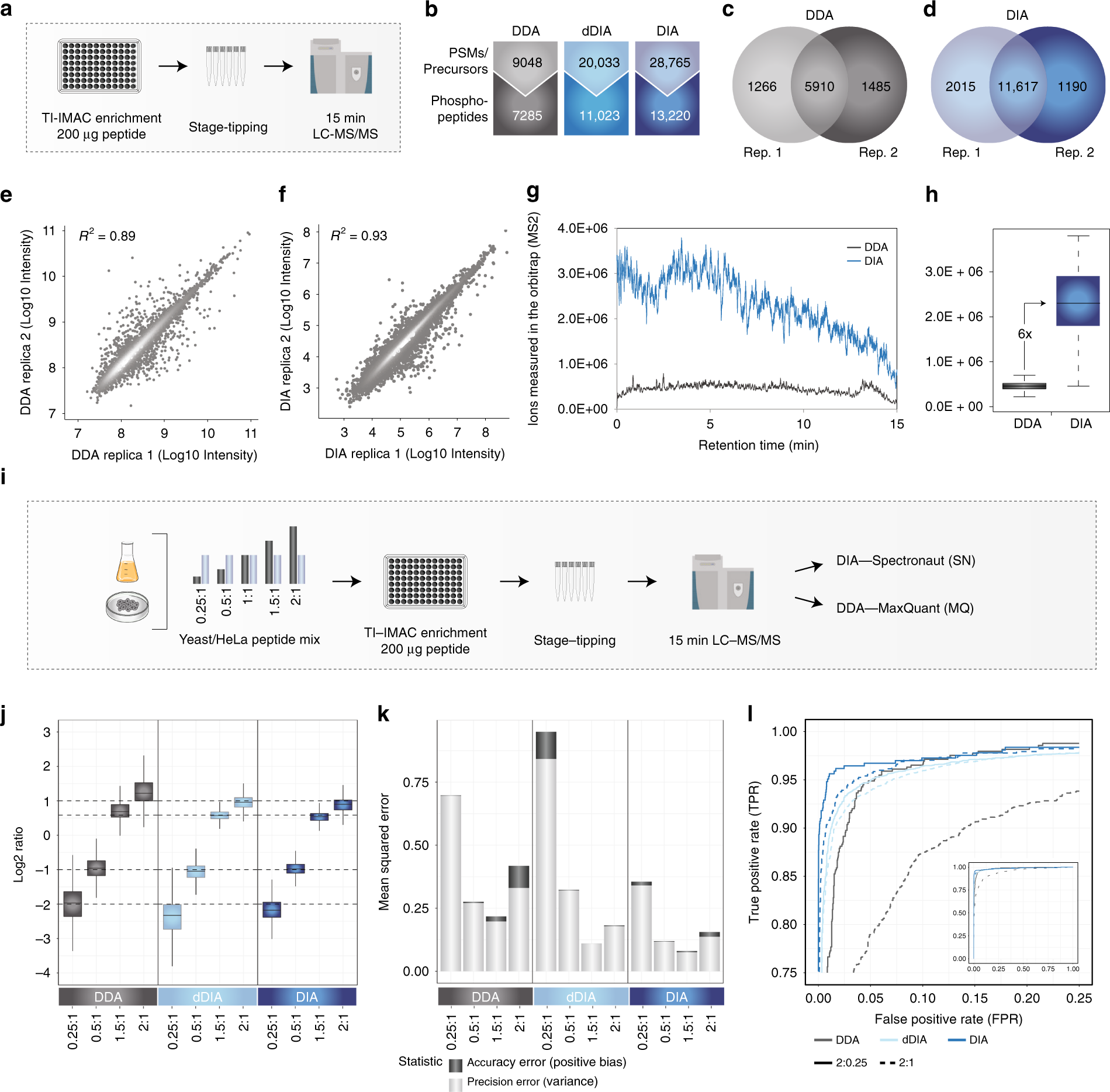
Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries | Nature Communications
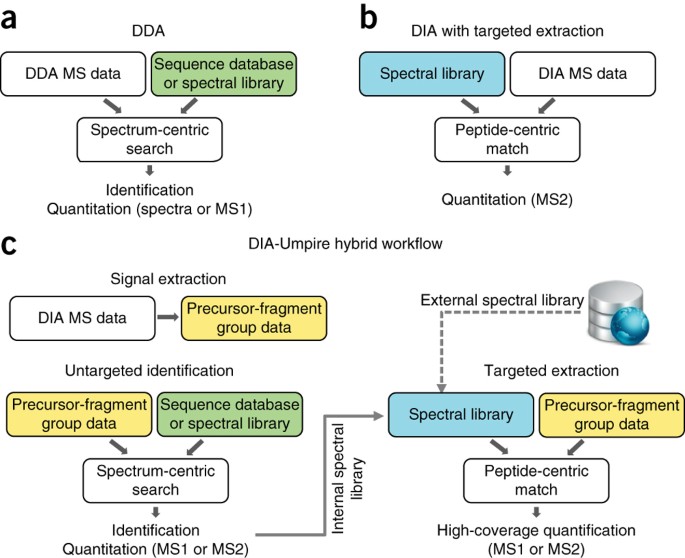
DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics | Nature Methods
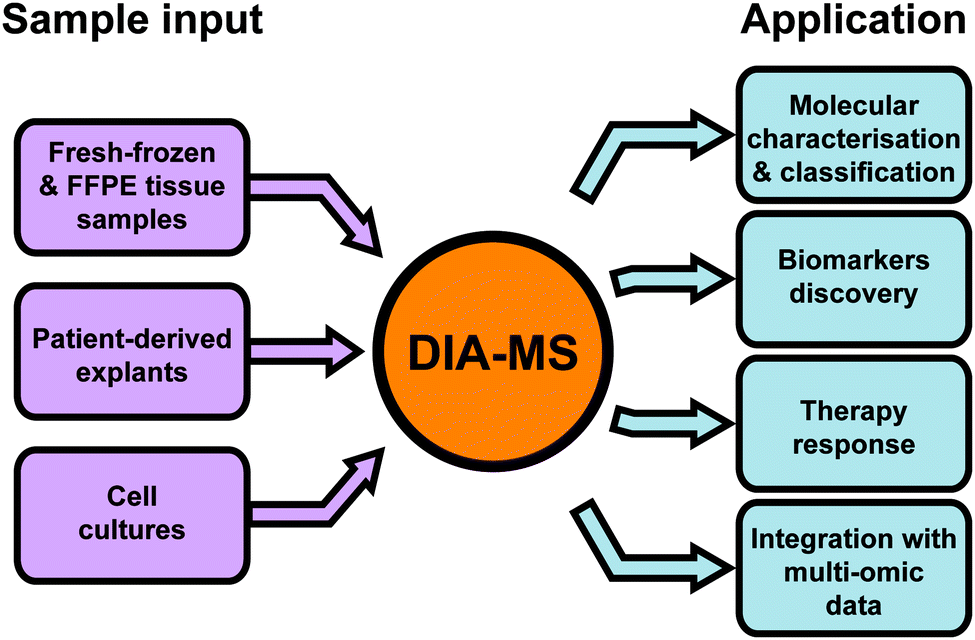
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H
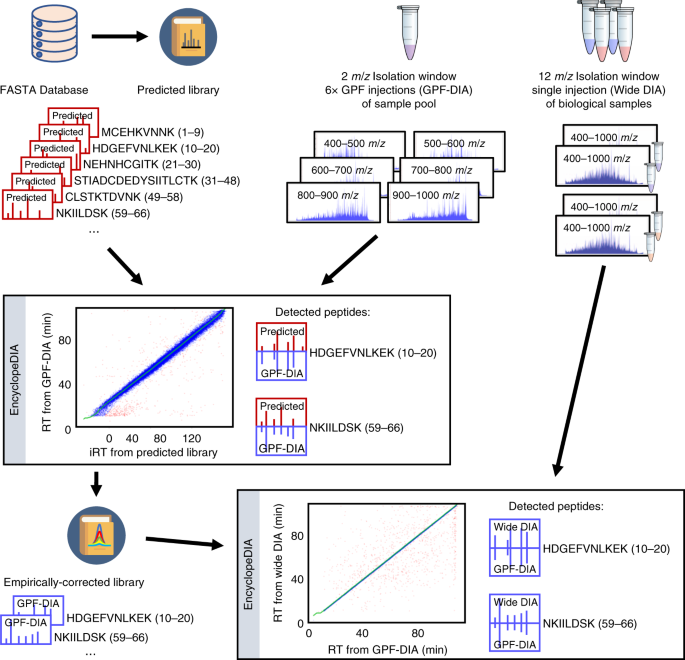
Generating high quality libraries for DIA MS with empirically corrected peptide predictions | Nature Communications
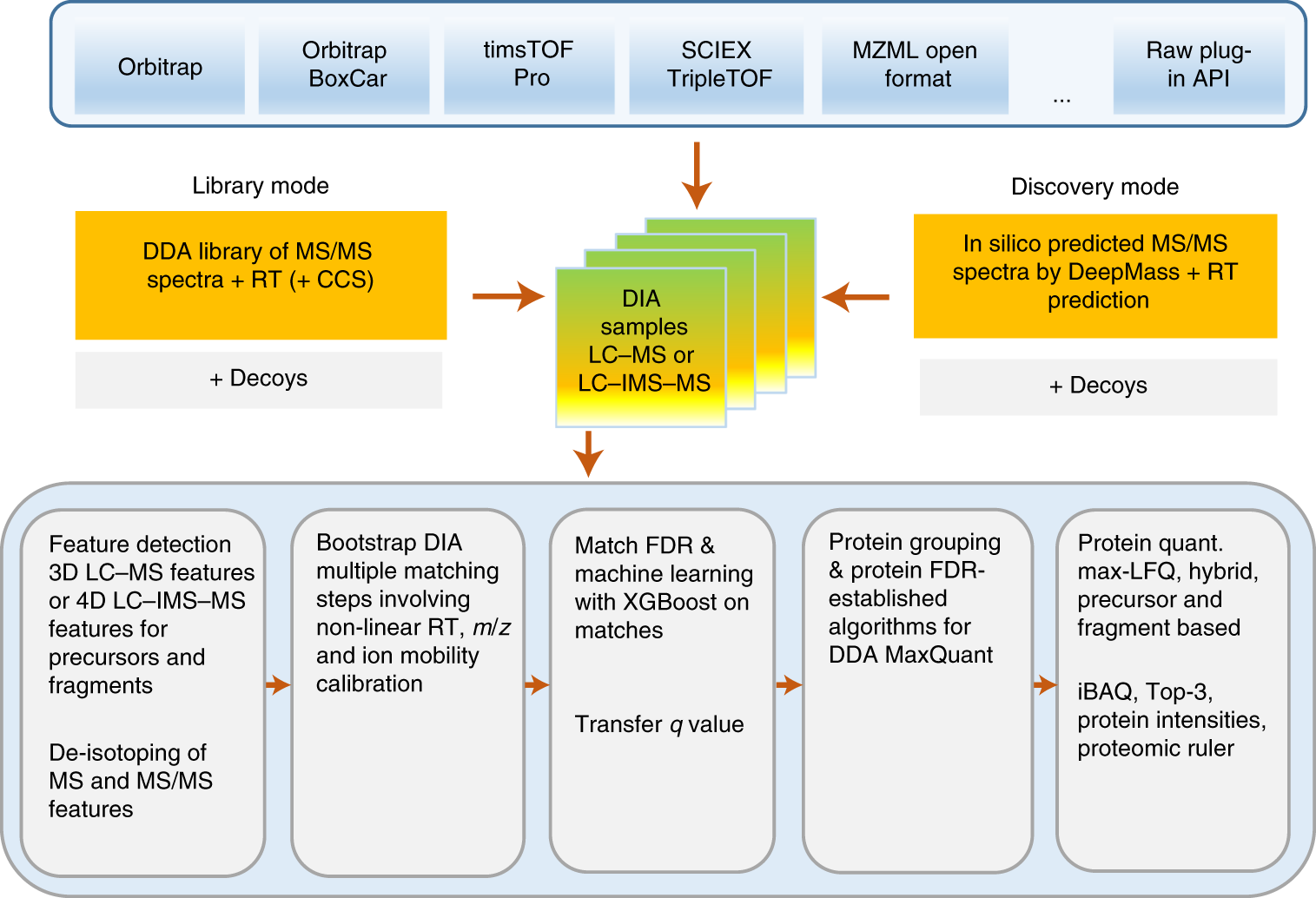
MaxDIA enables library-based and library-free data-independent acquisition proteomics | Nature Biotechnology

Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology | Nature Communications
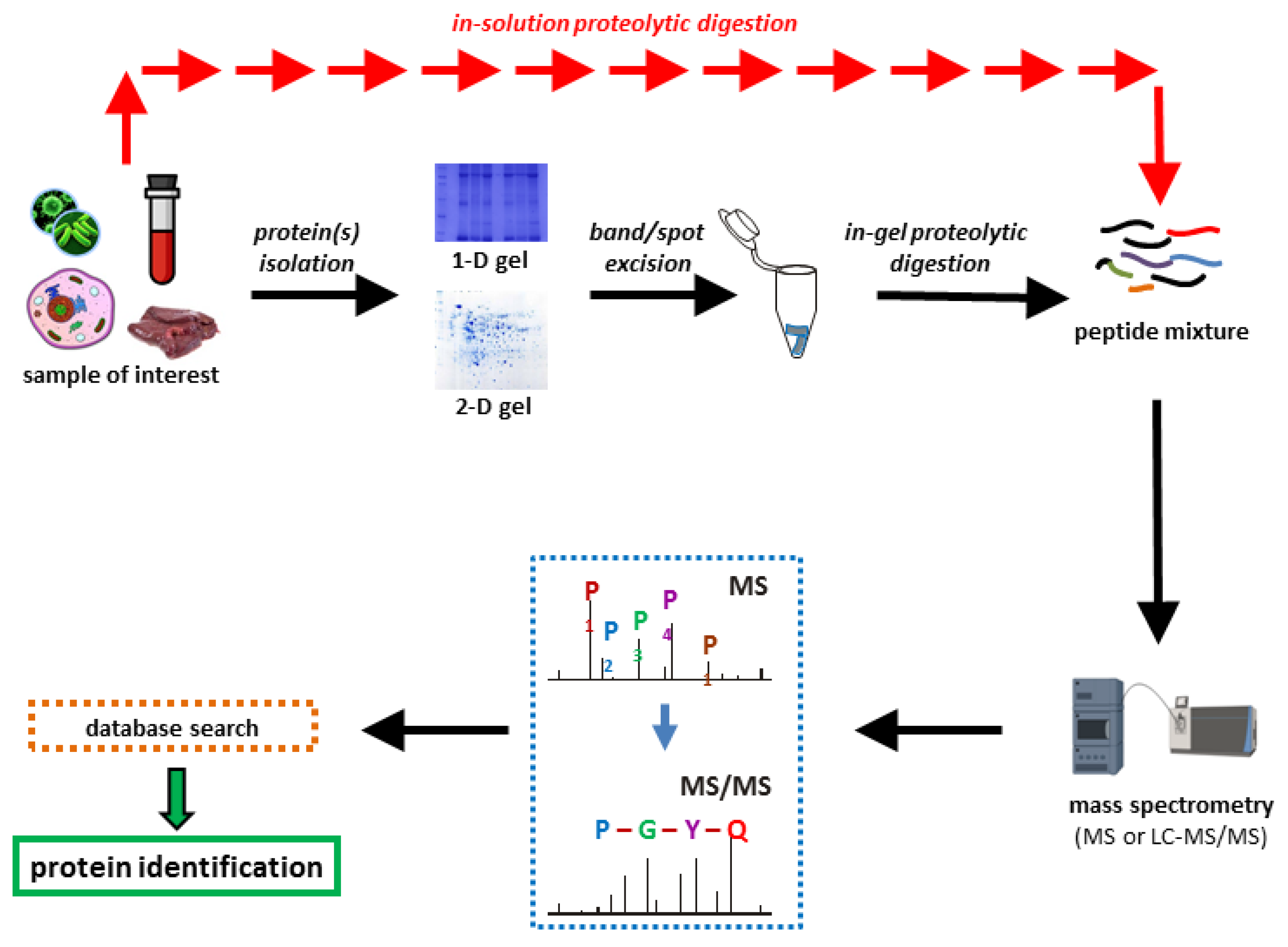
Proteomes | Free Full-Text | A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of This Field

Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics | Nature Communications

Technical comparison of DDA and different types of DIA in a biological... | Download Scientific Diagram

Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry | Nature Communications

Sensitive Immunopeptidomics by Leveraging Available Large-Scale Multi-HLA Spectral Libraries, Data-Independent Acquisition, and MS/MS Prediction - ScienceDirect
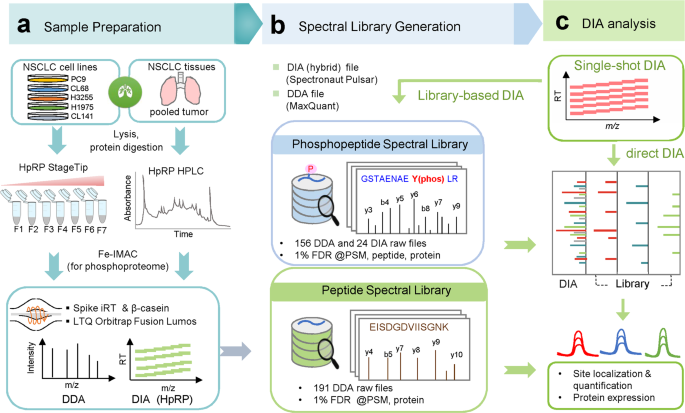
A data-independent acquisition-based global phosphoproteomics system enables deep profiling | Nature Communications